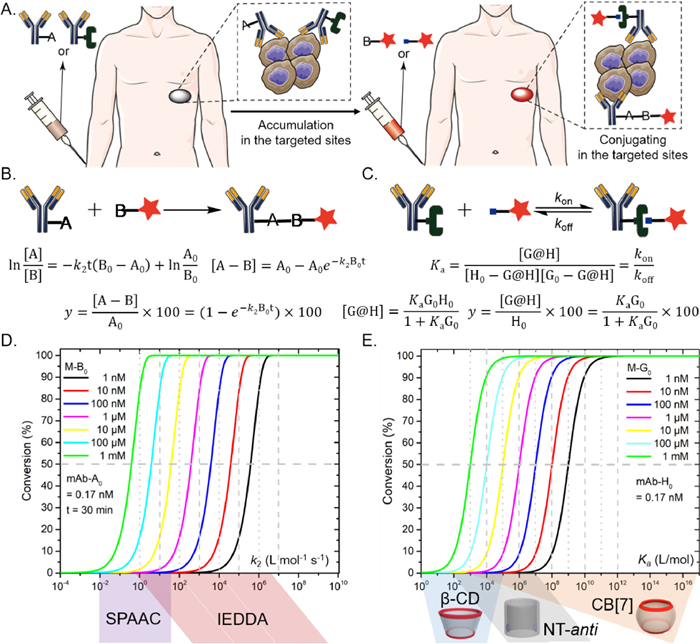
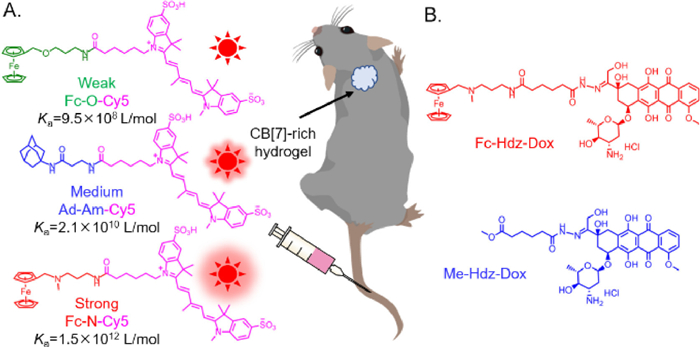
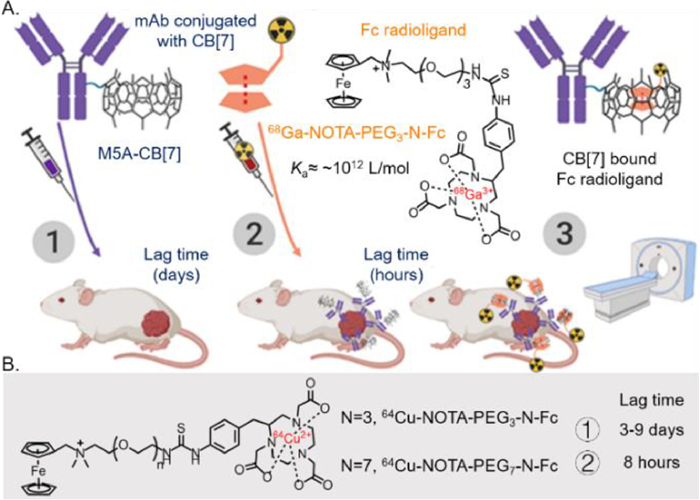
In physiological or pathological conditions, extracellular matrix (ECM) undergoes complex and diverse changes in structural and mechanical properties to support critical interactions with cells. Including stiffness, shear stress, stretching, ligand density, composition, and topography, have been confirmed to regulate cell behaviors. Particularly, tissue stiffening, an unique phenomenon of cancer progression, has attracted plentiful researches on specific cell behaviors in 2D planar systems [1-4]. However, as cells are surrounded by 3D ECM niches in vivo, the dimensionality shift may directly cause differences in ligand distribution, cell adhesion mode and cell stiffness-sensing, which would definitely affect cell behaviors [5,6]. For instance, in context of pathological diseases, cell metastasis is always accelerated by the increasing stiffness of traditional 2D substrates, while observations on 3D ECM mimics may be contradictory [7]. Thus, 3D ECM has been shown to be necessary to further explore the ability of artificial platforms, specifically in cell mechanical sensing.
To date, the fabrication of diverse 3D models has been achieved by many methods, such as cell spheroid culture, 3D bioprinting, tissue-engineering scaffolds with customizable properties, hydrogels, cell sheets, bioreactors with dynamic loading, and microfluidics [8]. Among them, microfluidics show the advantages of high throughput, stability and automation, making it possible to obtain a great quantity of samples with consistent spatial distribution and structures using low amounts of reagents [9-12]. In terms of materials, hydrogels combine the unique properties of softness, low immunogenicity and tunable mechanical characteristics [13-15]. Thus, monodisperse microgels produced with these elements not only offer exquisite control on homogeneous internal structures, but also shows the advantage in ECM physical features simulation [16]. Additionally, the limited size of microgels can avoid cells inactivation at internal positions caused by deficient oxygen and nutrients [17].
However, the microgels still present some limitations in the simulation of tumor stiffness due to their nature of high water content and flexibility [18]. To achieve high stiffness, simply increasing the weight concentration is not feasible, as it leads to a simultaneous change in porosity and viscosity, which brings out challenges including microflow shearing and microchip clogging [19]. Moreover, while the mechanical properties can be altered by the amount, type of crosslinking ions or methods, stiffness changes on orders-of-magnitude is hardly achieved [20]. To solve this problem, nanofibers have shown the prospect in simulation of increased collagenous fibrils in hardened tissue [21]. For example, Joshi et al. have successfully combined β-TCP incorporated PLA nanofibers with gelatin for biomimetic bone tissue engineering, the compressive stress was amplified about 8-fold [22]. It showed the nanofiber-hydrogel hybrids contained widely adjustable mechanical properties, which could fit the stiffness of some specific or diseased tissue.
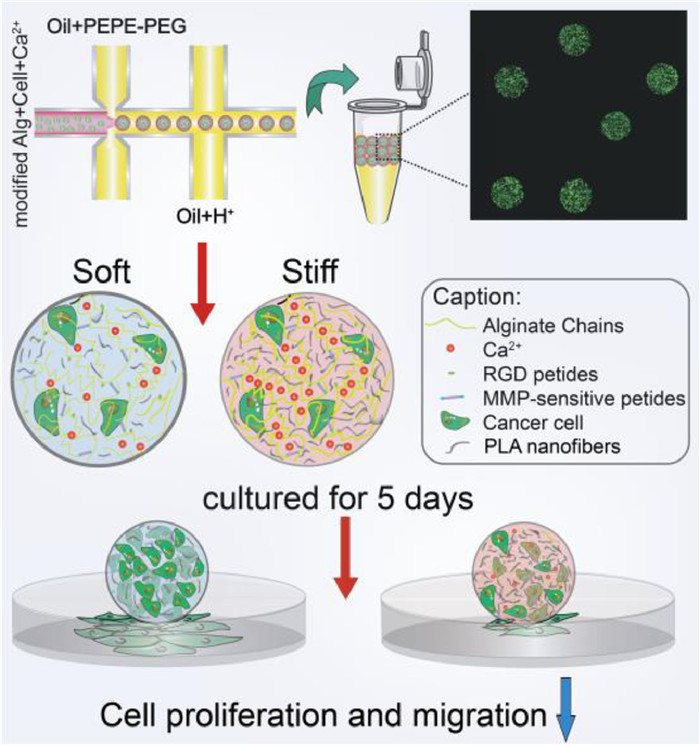
Herein, we present an innovative strategy to reinforce 3D microgel constructs by formulating composite hydrogel composed of modified alginate and different amount of micron-scale polylactide (PLA) nanofibers for solid carcinoma engineering (Scheme 1). The modified alginate with PVGLIG (proline-valine-glycine-leucine-isoleucine-glycine) and RGD (arginine-glycine-aspartate) peptides provided corresponding sites for cell migration and adhesion. By adding microscale PLA nanofibers uniformly by ultrasound, the composite microgels with increased stiffness showed the similar pore size, proving that we have created a 3D mechanical matrix platform with univariate variables. Following this strategy, solid breast tumors within the pathological breast stiffness range (2–20 kPa) were successfully simulated by using different composite gel ratios and two breast cancer cells with different invasive abilities, epithelial phenotype breast cancer cells MCF-7 and more aggressive mesenchymal SUM-159 [23]. Obviously, high proliferation and invasion ability of both cell types in soft matrix were observed. Furthermore, RT-qPCR analysis revealed the underlying changes at the genetic level. In this study, the proposed 3D microgel with homogeneous microstructure and adjustable stiffness effectively simulates the mechanical microenvironment of solid breast tumors, and demonstrates the advantages in-vitro simulation and tissue engineering.
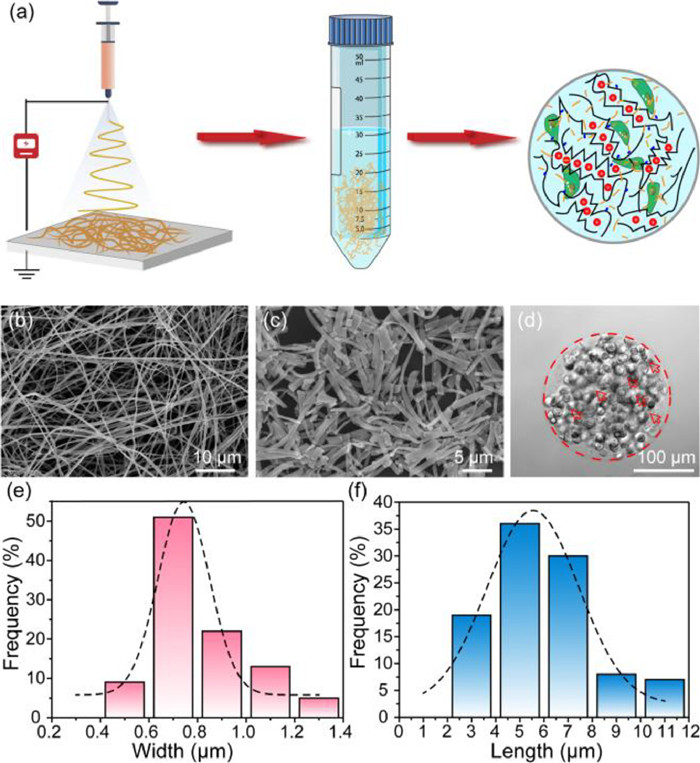
Firstly, PLA nanofibers with well-controlled diameters were fabricated by electrospinning process (Fig. 1a). Prior to use, morphology of nanofibers should be thoroughly characterized. As shown in Fig. 1b, the SEM micrograph of PLA fibers had random orientation and uniform width after electrospinning, and the average width was about 0.52 ± 0.12 µm, which fitted the Gaussian distribution (Fig. S1 in Supporting information). However, the fibers produced by electrospinning directly are always with irregular length in the tens or hundreds of microns, which might clog the outlets of microchips and hinder the uniform generation of micro-scale microgels. The homogeneous PLA nanofibers that shorter than the diameter of microgels are urgently needed to avoid the large agglomeration. Thus, the electrospun PLA nanofibers were aminolyzed in 5% ethylenediamine to obtain the micro-short scale, in order to disperse it uniformly in hydrogels [24]. And relevant experiments were optimized to discovery the influence of aminolysis time on the degree of fiber breakage. In Fig. S2 (Supporting information), PLA nanofibers became shorter and thicker as the increasing time of ammonolysis. For the 3 h ammonolysis (Fig. 1c), the PLA nanofibers were decomposed into micrometer-scale lengths, which had been suitable for making uniformly sized microgel spheres (Fig. 1d). Therefore, the 3 h ammonolysis PLA fibers with an average width and length of 0.8062 ± 0.1966 µm (Fig. 1e) and 5.9048 ± 2.1419 µm (Fig. 1f) could be employed in the composite hydrogels. And to guarantee the composite hydrogels biocompatible and stability, RGD and PVGLIG (MMP sensitive sequence) were introduced efficiently on alginate in response to cell adhesion and migration (Fig. S3 in Supporting information).
Next, we fabricated the microfluidic device to produce cell-loaded alginate/PLA hydrogel spheroids by flow-focusing (Fig. S4 in Supporting information) [25]. And the height and length of all microchannels were around 100 µm and 200 µm. Subsequently, the conditions of the continuous phases were optimized by fixing the velocity of dispersed phase. As shown in Videos S1–S4 (Supporting information), when the flow rate of the continuous phase was speeder than 600 µL/h, the dispersed phase at a rate of 40 µL/h could be cut into microgel spheres. To conform to the size requirement (~200 µm), we ultimately chose two continuous phases at the flow rate of 600 µL/h. The obtained microgels were collected in 20% 1H, 1H, 2H, 2H-perfluoro-1-octanol for demulsification.
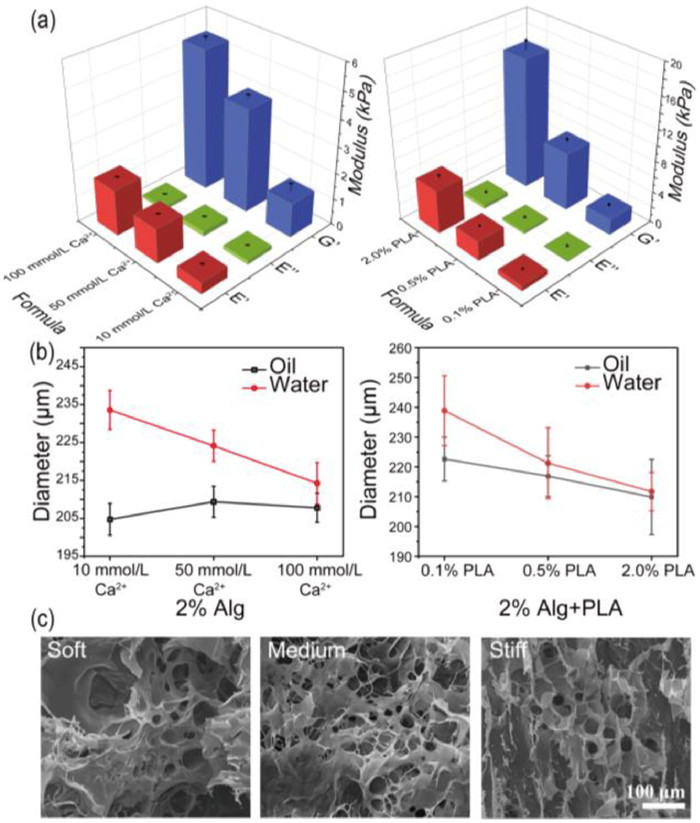
Subsequently, to simulate varying degrees of stiffening in breast cancer, different amounts of PLA nanofibers added in the compound gels should be explored. And the three groups in Table S1 (Supporting information) were finally selected. As shown in Fig. 2a and Fig. S5 (Supporting information), the viscosity modulus of 2.0% alginate with different concentrations of Ca2+ were far below the elastic modulus, indicating the Young's modulus was mainly affected by the modulus of elasticity. After the corresponding experimental groups that added different amounts of PLA, the viscous modulus of the composite hydrogel still kept the same trend, while the elastic modulus was greatly increased, thereby raising the overall stiffness about 1.54, 1.88, 3.17 times, up to 2.02 ± 0.25 kPa, 7.45 ± 0.49 kPa and 16.76 ± 1.18 kPa, respectively. Meanwhile, the swelling phenomenon during long-term culture also is a crucial feature, and the swelling ratio which reflected microgels cross-linking density was determined by the hydrodynamic diameter calculation of the microgel formulation in oil or water. As shown in Fig. 2b and Fig. S6 (Supporting information), the sizes of pure alginate groups are swelled from 204.73 ± 4.19 µm, 209.39 ± 4.10 µm, 207.80 ± 3.84 µm to 233.57 ± 5.10 µm, 224.14 ± 4.51 µm, 214.25 ± 5.49 µm. Microgels cross-linking with only 10 mmol/L Ca2+ swelled the most remarkable with 14.09% increasing. After adding PLA, the stability of the gel was further improved. The particle sizes are swelled from 222.64 ± 7.34 µm, 216.89 ± 6.82 µm, 209.89 ± 12.65 µm to 238.88 ± 11.73 µm, 221.25 ± 11.87 µm, 211.72 ± 6.41 µm, showing the swelling rates of three gel formulations were reduced to 7.29%, 2.01%, and 0.87%, respectively. Simultaneously, SEM images in Fig. 2c and Fig. S7 (Supporting information) demonstrated the homogeneity of pore size of three formulas, which were about 23.21 ± 7.71 µm, 24.47 ± 4.50 µm and 20.18 ± 8.50 µm, respectively. Statistical results showed that changing the cross-linking state within a certain range Ca2+ concentration would not affect the pore size of gels obviously, thus avoiding the influence in nutrients or the metabolic waste transport. Besides, the distribution of PLA micron nanofibers and the morphology of the cells encapsulated in different gel could be clearly observed from SEM images in Fig. S8 (Supporting information).
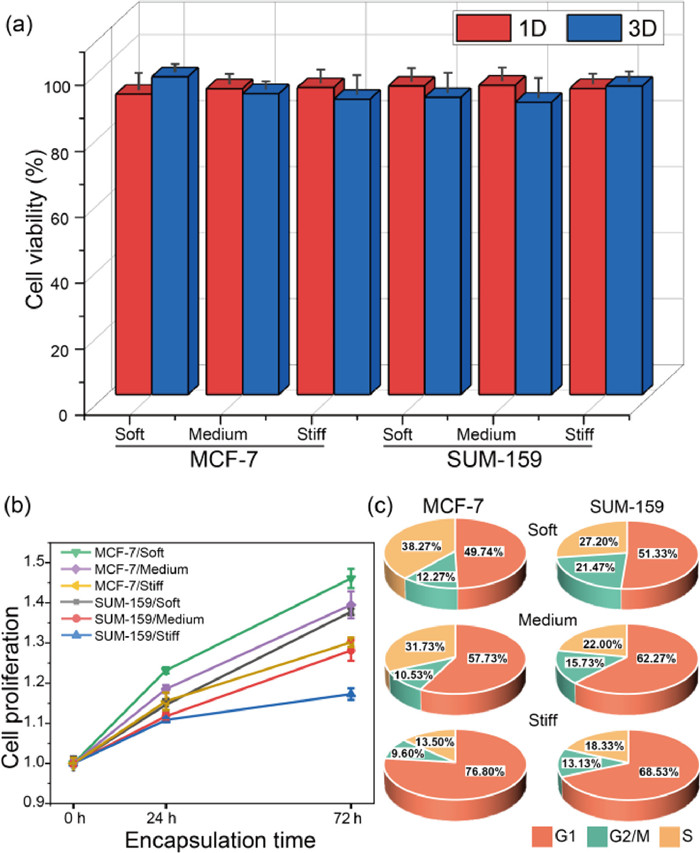
We proceeded to explore the role of stiffness on cell viability and proliferation in tumor microenvironments simulated by microgels. As shown in Fig. S9 (Supporting information), activity of MCF-7 and SUM-159 cells maintained no significant difference for all three conditions, similar results were obtained for other cell lines as well (Fig. S10 in Supporting information). And through counting and quantitative analysis of encapsulated cells of 1–3 days cultivation in Fig. 3a, the viability was at the range of 88.63% to 96.37%, which proved the high cytocompatible of nanofibers-composited hydrogels. Due to the modification of RGD and PVGLIG motifs, the hydrogel provided suitable sites for cell-matrix intergration and further improved cell growth and proliferation. In Fig. 3b, we estimated cell numbers on day 0, 1, and 3 by CCK8 assay and defined the fold change in proliferation by normalizing to the day 0 data. Two kinds of cells in the soft (2.02 kPa) and medium-stiffness (7.45 kPa) microenvironments showed significantly higher growth than the stiff (16.76 kPa) condition in 72 h. Subsequently, cell cycles were detected by flow cytometry. In Fig. 3c, the cells grown in the soft environments showed more distribution in the G2/M period, indicating the higher proliferation ability, which were consistent with the CCK8 results. In conclusion, both MCF-7 and SUM-159 cells tends to proliferate in a softer 3D microenvironment.
It is well known that breast cancer cells MCF-7 and SUM-159, exhibiting epithelial phenotype and mesenchymal phenotype, displayed with low and high metastatic potential in 2D plane, respectively (Fig. S11 in Supporting information). To study whether cell migration viability is highly correlated with 3D ECM stiffness, we encapsulated cells in 200 µm microgels and observed cell migrated from the inside of the gel to the outer space in 5 days.
Furthermore, we sought to further testify motility of encapsulated cells by immunofluorescence staining of N-cadherin and E-cadherin. As shown in Fig. S13 (Supporting information), cultured for 72 h, both N-cadherin and E-cadherin were observed in two type of cells. And the expression of E-cadherin was slightly lower in cells within the soft microgels, indicating the weakened intercellular cohesion. Thus, cells easily dispersed and infiltrated to the periphery. Obviously, with the increasing ECM stiffness, the decrease of N-cadherin expression level suggested the weakening of cell migration and invasion ability as well (Fig. 4d). Instead, the expression of epithelial markers E-cadherin in MCF-7 and mesenchymal markers N-cadherin in SUM-159 were more deeply affected by ECM stiffness, respectively, which was determined by cell characteristics. In summary, cell migration became tough in stiff 3D microenvironment. Therefore, we speculated that cells need time to find or pave a way to pass through the ECM with more physical constraints.
After demonstrating the association between migration and ECM stiffness in encapsulated cells, the changes induced by environmental confinement should be more visualized by gene expression. Matrix metalloproteinases (MMPs) are known to play an important role in developmental and homeostatic remodeling of ECM by mediating the degradation of ECM proteins, while tissue inhibitor of metalloproteinases (TIMPs) can adjust the MMPs expression [26]. From the point of view of migration ability, those genes were selected for further mRNA expression level analysis, and the detail of sequences were shown in Table S2. As shown in Fig. 4e, the expression of MMP2, MMP13, MMP14 and TIMP2 in both cells were significantly upward modulation within stiffer microgels. Meanwhile, the expression of those genes in 2D plane were downward than 3D condition, except the MMP13 in MCF-7 cells. Subsequently, MMP9 and TIMP1 of two kinds of breast cells were significantly down-regulated in stiff 3D microenvironment, and only MMP9 of SUM-159 expressed more in 3D ECM than 2D cultured. These observations were consistent with the known study that MMP2 and MMP9 could be repressed by TIMP1 and TIMP2, respectively. In addition, MMP3 and MMP8 of MCF-7 were significantly up-regulated with ECM stiffness, while the expression of those two genes demonstrated opposite in SUM-159. As for TIMP3, the expression was rapid declined in MCF-7 but raised in SUM-159 as stiffness changed. Those gene expression analysis revealed a complex and sensitive feedback network of cell migration upon 3D mechanical stimulation, which may provide some insights into solid tumor research and therapy.
In summary, we employed a composite hydrogel of PLA nanofibers and modified alginate with different concentrations of Ca2+, which can typically reinforce the mechanical properties of microgels with similar pore size. And we also investigated the effects of 3D microenvironment stiffness on the cell proliferation, viability, migration, and related gene expression of two types of breast cancer cells. Highly biocompatible and throughput microgels propose a new approach for the construction of solid tumors and observation of tumor migration. We envision that this study will provide an essential foundation for future investigations on ECM mechanisms in breast cancer and spark new approaches for therapy.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
This work was supported by the National Natural Science Foundation of China (Nos. 22034005, 81973569, and 21621003).
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2023.108319.
-
[1]
J.A. Prescher, C.R. Bertozzi, Nat. Chem. Biol. 1 (2005) 13–21.
doi: 10.1038/nchembio0605-13
-
[2]
E. Saxon, C.R. Bertozzi, Science 287 (2000) 2007–2010.
doi: 10.1126/science.287.5460.2007
-
[3]
H.C. Hang, C. Yu, D.L. Kato, C.R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A. 100 (2003) 14846–14851.
doi: 10.1073/pnas.2335201100
-
[4]
J.A. Prescher, D.H. Dube, C.R. Bertozzi, Nature 430 (2004) 873–877.
doi: 10.1038/nature02791
-
[5]
N.J. Agard, J.A. Prescher, C.R. Bertozzi, J. Am. Chem. Soc. 126 (2004) 15046–15047.
doi: 10.1021/ja044996f
-
[6]
E.M. Sletten, C.R. Bertozzi, Angew. Chem. Int. Ed. 48 (2009) 6974–6998.
doi: 10.1002/anie.200900942
-
[7]
E.M. Sletten, C.R. Bertozzi, Acc. Chem. Res. 44 (2011) 666–676.
doi: 10.1021/ar200148z
-
[8]
S.L. Scinto, D.A. Bilodeau, R. Hincapie, et al., Nat. Rev. Methods Prim. 1 (2021) 30.
doi: 10.1038/s43586-021-00028-z
-
[9]
C. Bertozzi, ACS Cent. Sci. 9 (2022) 558–559.
-
[10]
D.M. Patterson, L. Nazarova, J. Prescher, ACS Chem. Biol. 9 (2014) 592–605.
doi: 10.1021/cb400828a
-
[11]
M.L. Smeenk, J. Agramunt, K.M. Bonger, Curr. Opin. Chem. Biol. 60 (2021) 79–88.
doi: 10.1016/j.cbpa.2020.09.002
-
[12]
D. Wu, K. Yang, Z. Zhang, et al., Chem. Soc. Rev. 51 (2022) 1336–1376.
doi: 10.1039/D1CS00451D
-
[13]
A. Battigelli, B. Almeida, A. Shukla, Bioconjugate Chem. 33 (2022) 263–271.
doi: 10.1021/acs.bioconjchem.1c00564
-
[14]
L.I. Willems, W.A. van der Linden, N. Li, et al., Acc. Chem. Res. 44 (2011) 718–729.
doi: 10.1021/ar200125k
-
[15]
D.K. Nomura, M.M. Dix, B.F. Cravatt, Nat. Rev. Cancer 10 (2010) 630–638.
doi: 10.1038/nrc2901
-
[16]
K.S. Yang, G. Budin, C. Tassa, O. Kister, R. Weissleder, Angew. Chem. Int. Ed. 52 (2013) 10593–10597.
doi: 10.1002/anie.201304096
-
[17]
A. Rutkowska, D.W. Thomson, J. Vappiani, et al., ACS Chem. Biol. 11 (2016) 2541–2550.
doi: 10.1021/acschembio.6b00346
-
[18]
N.K. Devaraj, R. Weissleder, S.A. Hilderbrand, Bioconjug. Chem. 19 (2008) 2297–2299.
doi: 10.1021/bc8004446
-
[19]
N.K. Devaraj, S. Hilderbrand, R. Upadhyay, R. Mazitschek, R. Weissleder, Angew. Chem. Int. Ed. 49 (2010) 2869–2872.
doi: 10.1002/anie.200906120
-
[20]
H. Wu, J. Yang, J. Šečkutė, N.K. Devaraj, Angew. Chem. Int. Ed. 53 (2014) 5805–5809.
doi: 10.1002/anie.201400135
-
[21]
J. Yang, J. Seckute, C.M. Cole, N.K. Devaraj, Angew. Chem. Int. Ed. 51 (2012) 7476–7479.
doi: 10.1002/anie.201202122
-
[22]
Z.J. Gartner, C.R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 4606–4610.
doi: 10.1073/pnas.0900717106
-
[23]
G.A. Hudalla, W.L. Murphy, Langmuir 25 (2009) 5737–5746.
doi: 10.1021/la804077t
-
[24]
C.H. Kim, J. Axup, A. Dubrovska, et al., J. Am. Chem. Soc. 134 (2012) 9918–9921.
doi: 10.1021/ja303904e
-
[25]
Z.Y. Guan, C.Y. Wu, J.T. Wu, et al., ACS Appl. Mater. Interfaces 8 (2016) 13812–13818.
doi: 10.1021/acsami.6b03908
-
[26]
H. Koo, S.K. Hahn, S.H. Yun, Bioconjug. Chem. 27 (2016) 2601–2604.
doi: 10.1021/acs.bioconjchem.6b00546
-
[27]
H. Koo, M. Choi, E. Kim, et al., Small 11 (2015) 6458–6466.
doi: 10.1002/smll.201502972
-
[28]
E. Kim, H. Koo, Chem. Sci. 10 (2019) 7835–7851.
doi: 10.1039/C9SC03368H
-
[29]
N.K. Devaraj, ACS Cent. Sci. 4 (2018) 952–959.
doi: 10.1021/acscentsci.8b00251
-
[30]
R.E. Bird, S.A. Lemmel, X. Yu, Q.A. Zhou, Bioconjugate Chem. 32 (2021) 2457–2479.
doi: 10.1021/acs.bioconjchem.1c00461
-
[31]
R. Rossin, M.S. Robillard, Curr. Opin. Chem. Biol. 21 (2014) 161–169.
doi: 10.1016/j.cbpa.2014.07.023
-
[32]
C.L. Schreiber, B.D. Smith, Nat. Rev. Chem. 3 (2019) 393–400.
doi: 10.1038/s41570-019-0095-1
-
[33]
E.J.L. Stéen, P.E. Edem, K. Nørregaard, et al., Biomaterials 179 (2018) 209–245.
doi: 10.1016/j.biomaterials.2018.06.021
-
[34]
G. Liu, E.A. Wold, J. Zhou, Curr. Top. Med. Chem. 19 (2019) 892–897.
doi: 10.2174/1568026619666190510091921
-
[35]
W. Yi, P. Xiao, X. Liu, et al., Sig. Transduct. Target. Ther. 7 (2022) 386.
doi: 10.1038/s41392-022-01250-1
-
[36]
M. Handula, K.T. Chen, Y. Seimbille, Molecules 26 (2021) 4640.
doi: 10.3390/molecules26154640
-
[37]
T. Liang, Z. Chen, H. Li, Z. Gu, Trends Chem. 4 (2022) 157–168.
doi: 10.1016/j.trechm.2021.11.008
-
[38]
B.L. Oliveira, Z. Guo, G.J.L. Bernardes, Chem. Soc. Rev. 46 (2017) 4895–4950.
doi: 10.1039/C7CS00184C
-
[39]
R. Zhang, J. Gao, G. Zhao, et al., Nanoscale 15 (2023) 461–469.
doi: 10.1039/D2NR06056F
-
[40]
R. Rossin, S.M. van den Bosch, W. ten Hoeve, et al., Bioconjugate Chem. 24 (2013) 1210–1217.
doi: 10.1021/bc400153y
-
[41]
R. Selvaraj, J.M. Fox, Curr. Opin. Chem. Biol. 17 (2013) 753–760.
doi: 10.1016/j.cbpa.2013.07.031
-
[42]
K.N. Houk, A.G. Leach, S.P. Kim, X. Zhang, Angew. Chem. Int. Ed. 42 (2003) 4872–4897.
doi: 10.1002/anie.200200565
-
[43]
A. Rondon, F. Degoul, Bioconjug. Chem. 31 (2020) 159–173.
doi: 10.1021/acs.bioconjchem.9b00761
-
[44]
R.J. McMahon, Avidin-Biotin Interactions: Methods and Applications, Humana Press, Totowa, NJ, 2008.
-
[45]
www.clinicaltrials.gov/ct2/show/study/NCT00988715.
-
[46]
G. Yu, K. Jie, F. Huang, Chem. Rev. 115 (2015) 7240–7303.
doi: 10.1021/cr5005315
-
[47]
X. Ma, Y. Zhao, Chem. Rev. 115 (2015) 7794–7839.
doi: 10.1021/cr500392w
-
[48]
M.A. Beatty, F. Hof, Chem. Soc. Rev. 50 (2021) 4812–4832.
doi: 10.1039/D0CS00495B
-
[49]
J. Zhou, G. Yu, F. Huang, Chem. Soc. Rev. 46 (2017) 7021–7053.
doi: 10.1039/C6CS00898D
-
[50]
W.C. Geng, J.L. Sessler, D.S. Guo, Chem. Soc. Rev. 49 (2020) 2303–2315.
doi: 10.1039/C9CS00622B
-
[51]
L. Taiariol, C. Chaix, C. Farre, E. Moreau, Chem. Rev. 122 (2022) 340–384.
doi: 10.1021/acs.chemrev.1c00484
-
[52]
M. Altai, R. Membreno, B. Cook, V. Tolmachev, B.M. Zeglis, J. Nucl. Med. 58 (2017) 1553–1559.
doi: 10.2967/jnumed.117.189944
-
[53]
F.C.J. van de Watering, M. Rijpkema, M. Robillard, W.J.G. Oyen, O.C. Boerman, Front. Med. 1 (2014) 44.
-
[54]
C.J. Addonizio, B.D. Gates, M.J. Webber, Bioconjugate Chem. 32 (2021) 1935–1946.
doi: 10.1021/acs.bioconjchem.1c00326
-
[55]
S.M. Cheal, S.K. Chung, B.A. Vaughn, N.K.V. Cheung, S.M. Larson, J. Nucl. Med. 63 (2022) 1302–1315.
doi: 10.2967/jnumed.121.262186
-
[56]
J. Murray, J. Sim, K. Oh, et al., Angew. Chem. Int. Ed. 56 (2017) 2395–2398.
doi: 10.1002/anie.201611894
-
[57]
S.K. Ghosh, A. Dhamija, Y.H. Ko, et al., J. Am. Chem. Soc. 141 (2019) 17503–17506.
doi: 10.1021/jacs.9b09639
-
[58]
G. Sung, S.Y. Lee, M.G. Kang, et al., Chem. Commun. 56 (2020) 1549–1552.
doi: 10.1039/C9CC09699J
-
[59]
W. Cao, X. Qin, T. Liu, ChemBioChem 22 (2021) 2914–2917.
doi: 10.1002/cbic.202100357
-
[60]
K.L. Kim, G. Sung, J. Sim, et al., Nat. Commun. 9 (2018) 1712.
doi: 10.1038/s41467-018-04161-4
-
[61]
J. An, S. Kim, A. Shrinidhi, et al., Nat. Biomed. Eng. 4 (2020) 1044–1052.
doi: 10.1038/s41551-020-0589-7
-
[62]
D.V.D.W. Kankanamalage, J.H.T. Tran, N. Beltrami, et al., J. Am. Chem. Soc. 144 (2022) 16502–16511.
doi: 10.1021/jacs.2c05726
-
[63]
W. Cao, X. Qin, Y. Wang, et al., Angew. Chem. Int. Ed. 60 (2021) 11196–11200.
doi: 10.1002/anie.202100916
-
[64]
A. Kataki-Anastasakou, S. Hernandez, E.M. Sletten, ACS Chem. Biol. 16 (2021) 2124–2129.
doi: 10.1021/acschembio.1c00494
-
[65]
R. Sasmal, N.D. Saha, M. Pahwa, et al., Anal. Chem. 90 (2018) 11305–11314.
doi: 10.1021/acs.analchem.8b01851
-
[66]
A. Som, M. Pahwa, S. Bawari, et al., Chem. Sci. 12 (2021) 5484–5494.
doi: 10.1039/D0SC06860H
-
[67]
M. Li, A. Lee, K.L. Kim, et al., Angew. Chem. Int. Ed. 57 (2018) 2120–2125.
doi: 10.1002/anie.201711629
-
[68]
A. Lee, M. Li, Y.H. Ko, et al., Chem. Commun. 57 (2021) 12179–12182.
doi: 10.1039/D1CC04779E
-
[69]
B.D. Gates, J.B. Vyletel, L. Zou, M.J. Webber, Bioconjug. Chem. 33 (2022) 2262–2268.
doi: 10.1021/acs.bioconjchem.2c00242
-
[70]
F. Huang, J. Liu, Y. Liu, Chem. Sci. 13 (2022) 8885–8894.
doi: 10.1039/D2SC02797F
-
[71]
H. Tang, D. Fuentealba, Y.H. Ko, et al., J. Am. Chem. Soc. 133 (2011) 20623–20633.
doi: 10.1021/ja209266x
-
[72]
A. Douhal, Chem. Rev. 104 (2004) 1955–1976.
doi: 10.1021/cr020669j
-
[73]
A.A. Epenetos, D. Snook, H. Durbin, P.M. Johnson, J. Taylor-Papadimitriou, Cancer Res. 46 (1986) 3183–3191.
-
[74]
G.G. Bornstein, AAPS J. 17 (2015) 525–534.
doi: 10.1208/s12248-015-9738-4
-
[75]
I. Mahmood, Antibodies 10 (2021) 20.
doi: 10.3390/antib10020020
-
[76]
Y. Matsuda, B.A. Mendelsohn, Chem. Pharm. Bull. 69 (2021) 976–983.
doi: 10.1248/cpb.c21-00258
-
[77]
S.Y. Wu, F.G. Wu, X. Chen, Adv. Mater. 34 (2022) 2109210.
doi: 10.1002/adma.202109210
-
[78]
X. Shu, K. Xu, D. Hou, C. Li, Isr. J. Chem. 58 (2018) 1230–1240.
doi: 10.1002/ijch.201800115
-
[79]
T. Ogoshi, T.-a. Yamagishi, Y. Nakamoto, Chem. Rev. 116 (2016) 7937–8002.
doi: 10.1021/acs.chemrev.5b00765
-
[80]
W. Liu, E.M. Peck, K.D. Hendzel, B.D. Smith, Org. Lett. 17 (2015) 5268–5271.
doi: 10.1021/acs.orglett.5b02633
-
[81]
C.F.A. Gómez-Durán, W. Liu, M.L. Betancourt-Mendiola, B.D. Smith, J. Org. Chem. 82 (2017) 8334–8341.
doi: 10.1021/acs.joc.7b01486
-
[82]
M.V. Rekharsky, Y. Inoue, Chem. Rev. 98 (1998) 1875–1918.
doi: 10.1021/cr970015o
-
[83]
J. Lagona, P. Mukhopadhyay, S. Chakrabarti, L. Isaacs, Angew. Chem. Int. Ed. 44 (2005) 4844–4870.
doi: 10.1002/anie.200460675
-
[84]
A. Urbach, V. Ramalingam, Isr. J. Chem. 51 (2011) 664–678.
doi: 10.1002/ijch.201100035
-
[85]
K.I. Assaf, W.M. Nau, Chem. Soc. Rev. 44 (2015) 394–418.
doi: 10.1039/C4CS00273C
-
[86]
S.J. Barrow, S. Kasera, M.J. Rowland, J. del Barrio, O.A. Scherman, Chem. Rev. 115 (2015) 12320–12406.
doi: 10.1021/acs.chemrev.5b00341
-
[87]
H. Yao, H. Ke, X. Zhang, et al., J. Am. Chem. Soc. 140 (2018) 13466–13477.
doi: 10.1021/jacs.8b09157
-
[88]
L.L. Wang, Z. Chen, W.E. Liu, et al., J. Am. Chem. Soc. 139 (2017) 8436–8439.
doi: 10.1021/jacs.7b05021
-
[89]
L.P. Yang, X. Wang, H. Yao, W. Jiang, Acc. Chem. Res. 53 (2020) 198–208.
doi: 10.1021/acs.accounts.9b00415
-
[90]
Y.L. Ma, M. Quan, X.L. Lin, et al., CCS Chem. 3 (2021) 1078–1092.
doi: 10.31635/ccschem.020.202000288
-
[91]
Z. Liu, S.K.M. Nalluri, J.F. Stoddart, Chem. Soc. Rev. 46 (2017) 2459–2478.
doi: 10.1039/C7CS00185A
-
[92]
H. Yao, S.Y. Li, H. Zhang, et al., Chem. Commun. 59 (2023) 5411–5414.
doi: 10.1039/D2CC06622J
-
[93]
L. Cao, M. Šekutor, P.Y. Zavalij, et al., Angew. Chem. Int. Ed. 53 (2014) 988–993.
doi: 10.1002/anie.201309635
-
[94]
G. Hettiarachchi, D. Nguyen, J. Wu, et al., PLoS One 5 (2010) e10514.
doi: 10.1371/journal.pone.0010514
-
[95]
X. Zhang, X. Xu, S. Li, et al., Sci. Rep. 8 (2018) 8819.
doi: 10.1038/s41598-018-27206-6
-
[96]
M.G. Strebl, J. Yang, L. Isaacs, J.M. Hooker, Mol. Imaging 17 (2018), doi: 10.1177/1536012118799838.
doi: 10.1177/1536012118799838
-
[97]
S. Moghaddam, C. Yang, M. Rekharsky, et al., J. Am. Chem. Soc. 133 (2011) 3570–3581.
doi: 10.1021/ja109904u
-
[98]
L. Zou, A.S. Braegelman, M.J. Webber, ACS Cent. Sci. 5 (2019) 1035–1043.
doi: 10.1021/acscentsci.9b00195
-
[99]
M. Li, S. Kim, A. Lee, et al., ACS Appl. Mater. Interfaces 11 (2019) 43920–43927.
doi: 10.1021/acsami.9b16283
-
[100]
V.I.J. Jallinoja, B.D. Carney, M. Zhu, et al., Bioconjugate Chem. 32 (2021) 1554–1558.
doi: 10.1021/acs.bioconjchem.1c00280
-
[101]
V.I.J. Jallinoja, B.D. Carney, K. Bhatt, et al., Mol. Pharmaceut. 19 (2022) 2268–2278.
doi: 10.1021/acs.molpharmaceut.2c00102
-
[102]
A. Forero, P.L. Weiden, J.M. Vose, et al., Blood 104 (2004) 227–236.
-
[103]
S. Shen, A. Forero, A.F. LoBuglio, et al., J. Nucl. Med. 46 (2005) 642–651.
-
[104]
W. Wei, Z.T. Rosenkrans, J. Liu, et al., Chem. Rev. 120 (2020) 3787–3851.
doi: 10.1021/acs.chemrev.9b00738
-
[105]
V.I.J. Jallinoja, J.L. Houghton, J. Nucl. Med. 62 (2021) 1200–1206.
doi: 10.2967/jnumed.120.260687
-
[106]
H.C. Huang, S. Barua, G. Sharma, S.K. Dey, K. Rege, J. Control. Release 155 (2011) 344–357.
doi: 10.1016/j.jconrel.2011.06.004
-
[107]
N. Fernandes, C.F. Rodrigues, A.F. Moreira, I.J. Correia, Biomater. Sci. 8 (2020) 2990–3020.
doi: 10.1039/D0BM00222D
-
[108]
H. Zhu, B. Li, C.Y. Chan, et al., Adv. Drug Deliver. Rev. 192 (2023) 114644.
doi: 10.1016/j.addr.2022.114644
-
[109]
G. Yang, S.Z.F. Phua, A.K. Bindra, Y. Zhao, Adv. Mater. 31 (2019) 1805730.
doi: 10.1002/adma.201805730
-
[110]
Q. Cheng, L. Yue, J. Li, et al., Small 17 (2021) 2101332.
doi: 10.1002/smll.202101332
-
[111]
Q. Cheng, M. Xu, C. Sun, et al., Mater. Horiz. 9 (2022) 934–941.
doi: 10.1039/D1MH01813B
-
[112]
W. Li, K. Dong, H. Wang, et al., Biomaterials 217 (2019) 119310.
doi: 10.1016/j.biomaterials.2019.119310
-
[113]
G. Crini, Chem. Rev. 114 (2014) 10940–10975.
doi: 10.1021/cr500081p
-
[114]
Y. Chen, Y. Liu, Chem. Soc. Rev. 39 (2010) 495–505.
doi: 10.1039/B816354P
-
[115]
K.A. Connors, Chem. Rev. 97 (1997) 1325–1357.
doi: 10.1021/cr960371r
-
[116]
M.E. Brewster, T. Loftsson, Adv. Drug Deliver. Rev. 59 (2007) 645–666.
doi: 10.1016/j.addr.2007.05.012
-
[117]
S.V. Kurkov, T. Loftsson, Int. J. Pharmaceut. 453 (2013) 167–180.
doi: 10.1016/j.ijpharm.2012.06.055
-
[118]
T. Loftsson, M.E. Brewster, J. Pharm. Pharmacol. 62 (2010) 1607–1621.
doi: 10.1111/j.2042-7158.2010.01030.x
-
[119]
C. Kahle, U. Holzgrabe, Chirality 16 (2004) 509–515.
doi: 10.1002/chir.20068
-
[120]
Z. Yang, R. Breslow, Tetrahedron Lett. 38 (1997) 6171–6172.
doi: 10.1016/S0040-4039(97)01427-5
-
[121]
S.S. Agasti, M. Liong, C. Tassa, et al., Angew. Chem. Int. Ed. 51 (2012) 450–454.
doi: 10.1002/anie.201105670
-
[122]
E. Garin, Y. Rolland, S. Laffont, J. Edeline, Eur. J. Nucl. Med. Mol. I. 43 (2016) 559–575.
doi: 10.1007/s00259-015-3157-8
-
[123]
J.F. Prince, R. van Diepen, R. van Rooij, M.G. Lam, Nucl. Med. Commun. 37 (2016) 218–219.
doi: 10.1097/MNM.0000000000000424
-
[124]
S.J. Spa, M.M. Welling, M.N. van Oosterom, et al., Theranostics 8 (2018) 2377–2386.
doi: 10.7150/thno.23567
-
[125]
M.M. Welling, S.J. Spa, D.M. van Willigen, et al., J. Control. Release 293 (2019) 126–134.
doi: 10.1016/j.jconrel.2018.11.020
-
[126]
M.M. Welling, N. Duszenko, D.M. van Willigen, et al., Bioconjug. Chem. 32 (2021) 607–614.
doi: 10.1021/acs.bioconjchem.1c00061
-
[127]
M. Xu, J. Tao, Z. Wei, et al., Nano Today 43 (2022) 101450.
doi: 10.1016/j.nantod.2022.101450
-
[128]
B.J. Shorthill, C.T. Avetta, T.E. Glass, J. Am. Chem. Soc. 126 (2004) 12732–12733.
doi: 10.1021/ja047639d
-
[129]
G.B. Huang, S.H. Wang, H. Ke, L.P. Yang, W. Jiang, J. Am. Chem. Soc. 138 (2016) 14550–14553.
doi: 10.1021/jacs.6b09472
-
[130]
H. Ke, L.P. Yang, M. Xie, et al., Nat. Chem. 11 (2019) 470–477.
doi: 10.1038/s41557-019-0235-8
-
[131]
H. Chai, Z. Chen, S.H. Wang, et al., CCS Chem. 2 (2020) 440–452.
doi: 10.31635/ccschem.020.202000160
-
[132]
W.E. Liu, Z. Chen, L.P. Yang, H.Y. Au-Yeung, W. Jiang, Chem. Commun. 55 (2019) 9797–9800.
doi: 10.1039/C9CC04603H
-
[133]
L.M. Bai, H. Yao, L.P. Yang, W. Zhang, W. Jiang, Chin. Chem. Lett. 30 (2019) 881–884.
doi: 10.1016/j.cclet.2018.11.033
-
[134]
H. Yao, X. Wang, M. Xie, et al., Org. Biomol. Chem. 18 (2020) 1900–1909.
doi: 10.1039/D0OB00290A
-
[135]
Y.L. Ma, C. Sun, Z. Li, et al., CCS Chem. 4 (2022) 1977–1989.
doi: 10.31635/ccschem.021.202101178
-
[136]
S. Wilhelm, A.J. Tavares, Q. Dai, et al., Nat. Rev. Mater. 1 (2016) 16014.
doi: 10.1038/natrevmats.2016.14
-
[137]
Q. Dai, S. Wilhelm, D. Ding, et al., ACS Nano 12 (2018) 8423–8435.
doi: 10.1021/acsnano.8b03900
-
[138]
J. Liu, M. Li, Z. Luo, et al., Nano Today 15 (2017) 56–90.
doi: 10.1016/j.nantod.2017.06.010
-
[139]
K.T. Xenaki, S. Oliveira, P.M.P. van Bergen en Henegouwen, Front. Immunol. 8 (2017) 1287.
doi: 10.3389/fimmu.2017.01287
-
[140]
J.M. Mejia Oneto, I. Khan, L. Seebald, M. Royzen, ACS Cent. Sci. 2 (2016) 476–482.
doi: 10.1021/acscentsci.6b00150
-
[141]
Q. Jin, Z. Liu, Q. Chen, J. Control. Release 329 (2021) 882–893.
doi: 10.1016/j.jconrel.2020.10.019
-
[142]
Y.L. Liu, D. Chen, P. Shang, D.C. Yin, J. Control. Release 302 (2019) 90–104.
doi: 10.1016/j.jconrel.2019.03.031
-
[143]
M. Saadat, M.K.D. Manshadi, M. Mohammadi, et al., J. Control. Release 328 (2020) 776–791.
doi: 10.1016/j.jconrel.2020.09.017
-
[144]
L.A.L. Fliervoet, E. Mastrobattista, Adv. Drug Deliv. Rev. 106 (2016) 63–72.
doi: 10.1016/j.addr.2016.04.021
-
[145]
A.S. Timin, M.M. Litvak, D.A. Gorin, et al., Adv. Healthc. Mater. 7 (2018) 1700818.
doi: 10.1002/adhm.201700818
-
[146]
T. Zhang, R. Lin, H. Wu, X. Jiang, J. Gao, Adv. Drug Deliv. Rev. 185 (2022) 114300.
doi: 10.1016/j.addr.2022.114300
-
[147]
S. Abbina, E.M.J. Siren, H. Moon, J.N. Kizhakkedathu, ACS Biomater. Sci. Eng. 4 (2018) 3658–3677.
doi: 10.1021/acsbiomaterials.7b00514

 Login In
Login In





 DownLoad:
DownLoad:
 DownLoad:
DownLoad: