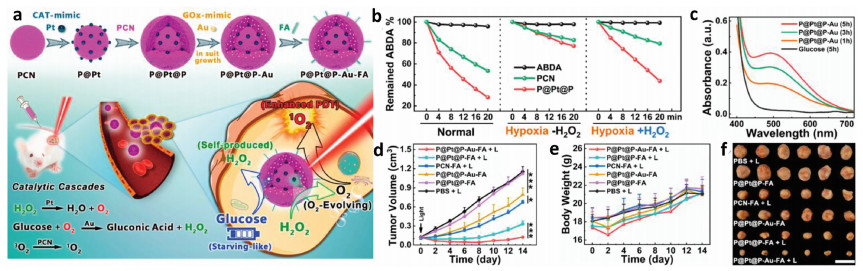
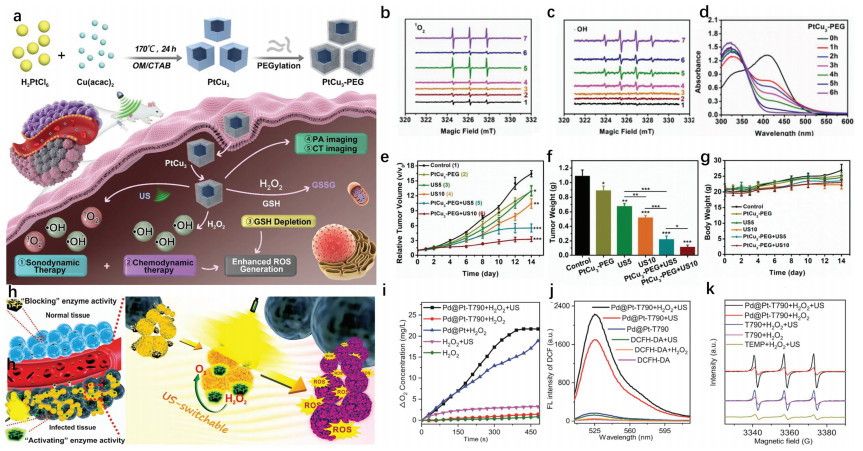
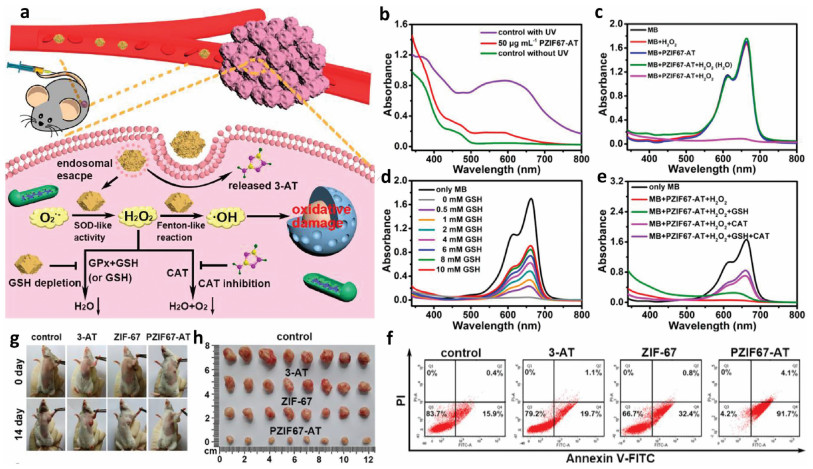
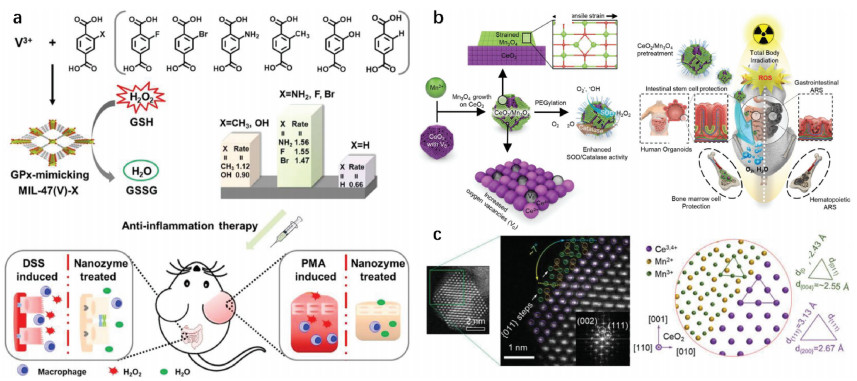
Adenosine triphosphate (ATP), known as "the energy currency of the cell", plays an important role in living cells, including protein synthesis, intracellular signal transduction and cell division [1,2]. The abnormal levels of intracellular ATP have been proved to be closely associated with a variety of diseases, such as cancer and Parkinson [3]. On the other hand, as a signal molecule, hydrogen sulfide (H2S) widely exists in heart, brain, liver and other major organs, and plays an irreplaceable role in physiological functions including neural regulation, apoptosis, insulin signal inhibition and blood pressure [4]. The normal concentration of H2S in cells is about 0.01–3 µmol/L [5]. Abnormal H2S level can lead to the disfunction of cell, which is related to many diseases, like cirrhosis and Alzheimer's disease [6]. Therefore, the detection of ATP and H2S level has important physiological and pathological significance.
Currently, fluorescent sensors for simultaneous detection dual or multiple chemical species in biology have attracted much attention [15–17]. Some multifunctional fluorescent sensors for ATP and another analyte, including H2O2, H2S, ONOO− and nitroreductase (NTR), were also documented [18–23]. Typically, the multi-analyte chemosensors were elegantly prepared by covalently linked multiple analyte recognition sites, including complexation and chemical reaction, with one or two flurophores in a single molecule. This strategy requires cumbersome chemical synthesis methods and is time-consuming and labor-intensive.
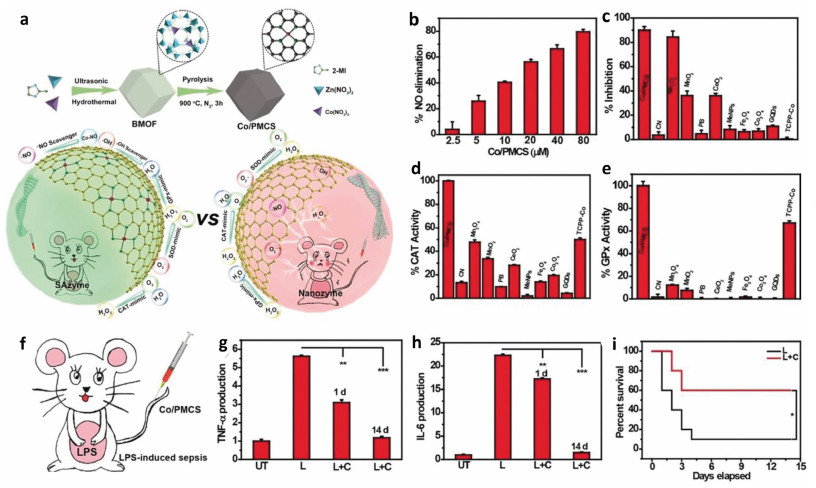
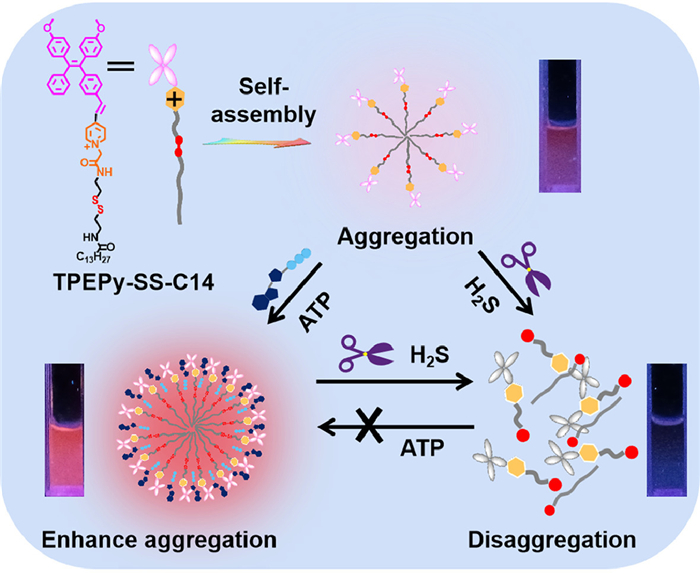
Aggregation-induced emission (AIE) fluorogens, also named as AIEgens, which have attracted much attention for their unique fluorescence emission properties [24]. The AIEgens show non- or weak emission in dilution solutions, but emit strongly at the aggregation state. Previously, we have reported imidazolium-functionalized tetraphenylethylenes, which have a good fluorescence turn-on sensing toward ATP for the complexation enhanced aggregation [25]. We envisioned that if the second recognition site such as the disulfide bond is rationally incorporated into this system, a dual functional probe which can detect ATP and another analyte will be obtained. In this context, herein, we design and synthesize a novel AIE bifunctional probe TPEPy-SS-C14 (Scheme 1), which can simultaneously detect ATP and H2S based on the aggregation/disaggregation mechanism. The probe TPEPy-SS-C14 is rationally designed as following: (1) The TPEPy unit makes probe a red AIE-based emission, (2) the pyridinium and amide groups are used as the ATP binding site via the electrostatic interactions and hydrogen bonding, (3) the disulfide bond is cleavable by H2S, and (4) the long alkyl chain endows probe a good amphiphilic property in aqueous solution. In addition, the probe locates mitochondria, and can detect ATP and hydrogen sulfide levels in living cells.
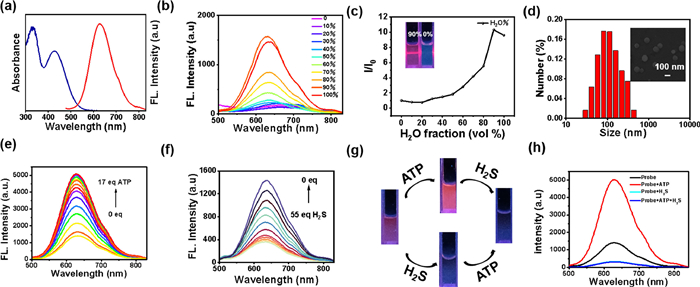
Firstly, the ultraviolet–visible (UV–vis) absorption and fluorescence spectra of TPEPy-SS-C14 (10 µmol/L) was investigated in aqueous solution. As shown in Fig. 1a, TPEPy-SS-C14 exhibits a red emission with the maximum peak at 630 nm and a low-energy absorption peak at 428 nm. The Stokes shift is 202 nm, which can prevent the interference effectively caused by self-absorption in the excitation process of biological imaging.
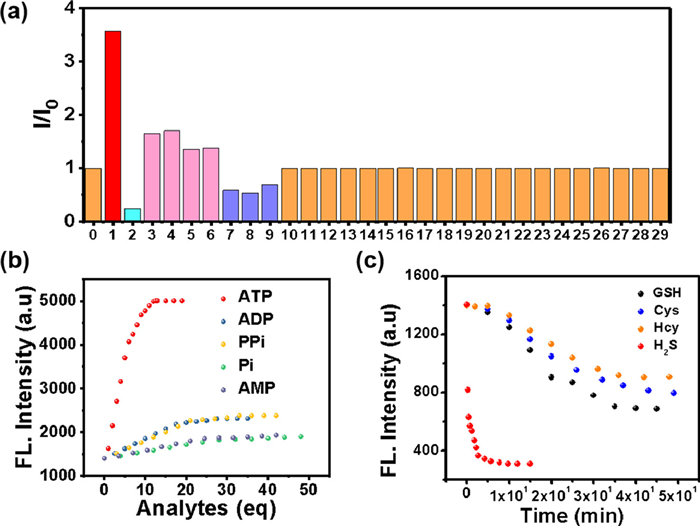
On the other hand, we find the addition of some thiols-containing amide acid, including glutathione (GSH), cysteine (Cys) and homocysteine (Hcy), can also leads to a little decrease of the fluorescence. However, the quench ratio by the thiols is much lower than that of H2S, indicating that TPEPy-SS-C14 can discriminate H2S from thiols and other analytes. The difference fluorescence response of TPEPy-SS-C14 toward H2S and thiols may be attributed their different reaction mechanism. For H2S, which mainly exists as HS− in physiological conditions, can effectively cleave the disulfide bond to release the fluorescent active species TPEPy-SSH [8–12]. However, the thiols reaction may give both the cleavage and the disulfide exchange products. We also studied the reaction mechanism of GSH with the probe (Fig. S17 in Supporting information), which captures not only the cleavage product TPEPy-SH, but also the disulfide exchange species TPEPy-SG. Regarding that TPEPy-SG is also amphiphily for bearing the hydrophilic glutathione group, it may exhibit self-assembly property with high emission. Therefore, the fluorescence is only slightly reduced after adding GSH and other thiols [26]. Furthermore, H2S shows stronger nucleophilicity than bio-thiols. We compared the time kinetics of H2S and other thiols with TPEPy-SS-C14 (Fig. 2c), and found that the fluorescence of hydrogen sulfide added decreased rapidly in a few seconds, and basically reached equilibrium in 5 min, while other thiols took nearly 50 min. So probe TPEPy-SS-C14 reacting with H2S is dominant under the complexed physiological conditions.
For really application, a good multifunctional probe should also behaves an excellent selectively in many competitive ions. Thus, the competitive experiments of TPEPy-SS-C14 toward ATP/H2S under the coexistence of many interference species were carried out (Fig. S18 in Supporting information). It was revealed that TPEPy-SS-C14 still displays a excellent fluorescence enhancement ability toward ATP in the presence of these analytes, except H2S. However, it can recognize H2S in the presence of the following 27 analytes, containing biothiols GSH, Cys and Hcy (Fig. S19 in Supporting information). In addition, the influences of pH experiments reveals that this probe can work in wide pH range of 6‒10, indicating the physiological applicability of the TPEPy-SS-C14 probe (Fig. S20 in Supporting information).
Based on the good selective competition and anti-interference ability of the probe in vitro, TPEPy-SS-C14 was used to recognize ATP and H2S in living cells. Firstly, the toxicity of TPEPy-SS-C14 on SMMC cells was conducted by standard MTT assay. As shown in Fig. S21 (Supporting information), TPEPy-SS-C14 possessed a low cytotoxicity at the test concentration and is appropriate for intracellular imaging. The subcellular location of TPEPy-SS-C14 was investigated by co localization experiment with Mito tracker green. TPEPy-SS-C14 overlaps well with Mito tracker green dye, with an overlap rate of 0.91 (Fig. S22 in Supporting information). These results show that TPEPy-SS-C14 has a good localization effect on mitochondria and provides the imaging ability of ATP and hydrogen sulfide in mitochondria.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
This work was supported by the National Nature Science Foundation of China (No. 22061028) and Jiangxi Provincial Natural Science Foundation (No. 20224ACB203012).
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2023.108925.
-
[1]
A.W. Dobson, Y. Xu, M.R. Kelley, et al., J. Biol. Chem. 275(2000) 37518-37523.
doi: 10.1074/jbc.M000831200
-
[2]
H. Beinert, D.E. Green, P. Hele, et al., Science 124(1956) 614-615.
doi: 10.1126/science.124.3223.614
-
[3]
C.N. Serhan, A. Jain, S. Marleau, et al., J. Immunol. 171(2003) 6856-6865.
doi: 10.4049/jimmunol.171.12.6856
-
[4]
S.C. Lu, Mol. Aspects Med. 30(2009) 42-59.
doi: 10.1016/j.mam.2008.05.005
-
[5]
D.M. Shih, L.J. Gu, Y.R. Xia, et al., Nature 394(1998) 284-287.
doi: 10.1038/28406
-
[6]
C. Mateo, J.M. Palomo, G.F. Lorente, et al., Enzyme Microb. Technol. 40(2007) 1451-1463.
doi: 10.1016/j.enzmictec.2007.01.018
-
[7]
M. Liang, X. Yan, Acc. Chem. Res. 52(2019) 2190-2200.
doi: 10.1021/acs.accounts.9b00140
-
[8]
F. Manea, F.B. Houillon, L. Pasquato, P. Scrimin, Angew. Chem. Int. Ed. 43(2004) 6165-6169.
doi: 10.1002/anie.200460649
-
[9]
L. Gao, J. Zhuang, L. Nie, et al., Nat. Nanotechnol. 2(2007) 577-583.
doi: 10.1038/nnano.2007.260
-
[10]
B. Liu, Z. Sun, P.J. Huang, J. Liu, J. Am. Chem. Soc. 137(2015) 1290-1295.
doi: 10.1021/ja511444e
-
[11]
F. Wang, Y. Zhang, Z. Liu, et al., Nanoscale 12(2020) 14465-14471.
doi: 10.1039/D0NR03217D
-
[12]
W. Yin, J. Yu, F. Lv, et al., ACS Nano 10(2016) 11000-11011.
doi: 10.1021/acsnano.6b05810
-
[13]
K. Zhang, M. Tu, W. Gao, et al., Nano Lett. 19(2019) 2812-2823.
doi: 10.1021/acs.nanolett.8b04729
-
[14]
C. Liu, J. Xing, O.U. Akakuru, et al., Nano Lett. 19(2019) 5674-5682.
doi: 10.1021/acs.nanolett.9b02253
-
[15]
R. Yan, S. Sun, J. Yang, et al., ACS Nano 13(2019) 11552-11560.
doi: 10.1021/acsnano.9b05075
-
[16]
M.P. Murphy, Biochem. J. 417(2009) 1-13.
doi: 10.1042/BJ20081386
-
[17]
C.C. Winterbourn, Nat. Chem. Biol. 4(2008) 278-286.
doi: 10.1038/nchembio.85
-
[18]
V.J. Thannickal, B.L. Fanburg, Am. J. Physiol. Lung Cell Mol. Physiol. 279(2000) L1005-L1028.
doi: 10.1152/ajplung.2000.279.6.L1005
-
[19]
S.S. Gill, N. Tuteja, Plant Physiol. Biochem. 48(2010) 909-930.
doi: 10.1016/j.plaphy.2010.08.016
-
[20]
H. Pelicano, D. Carney, P. Huang, Drug Resist. Updat. 7(2004) 97-110.
doi: 10.1016/j.drup.2004.01.004
-
[21]
E.R. Stadtman, Free Radical Res. 40(2006) 1250-1258.
doi: 10.1080/10715760600918142
-
[22]
K. Ito, A. Hirao, F. Arai, et al., Nat. Med. 12(2006) 446-451.
doi: 10.1038/nm1388
-
[23]
M. Mittal, M.R. Siddiqui, K. Tran, et al., Antioxid. Redox Signal. 20(2014) 1126-1167.
doi: 10.1089/ars.2012.5149
-
[24]
F. Giacco, M. Brownlee, Circul. Res. 107(2010) 1058-1070.
doi: 10.1161/CIRCRESAHA.110.223545
-
[25]
M. Brownlee, Diabetes 54(2005) 1615-1625.
doi: 10.2337/diabetes.54.6.1615
-
[26]
W.R. Markesbery, Free Radical Biol. Med. 23(1997) 134-147.
doi: 10.1016/S0891-5849(96)00629-6
-
[27]
K. Apel, H. Hirt, Annu. Rev. Plant Biol. 55(2004) 373-399.
doi: 10.1146/annurev.arplant.55.031903.141701
-
[28]
Z. Zhou, J. Song, L. Nie, X. Chen, Chem. Soc. Rev. 45(2016) 6597-6626.
doi: 10.1039/C6CS00271D
-
[29]
X. Qian, Y. Zheng, Y. Chen, Adv. Mater. 28(2016) 8097-8129.
doi: 10.1002/adma.201602012
-
[30]
S. Wang, G. Yu, Z. Wang, et al., Angew. Chem. Int. Ed. 58(2019) 14758-14763.
doi: 10.1002/anie.201908997
-
[31]
P. Sonveaux, Oncotarget 8(2017) 35482-35483.
doi: 10.18632/oncotarget.16613
-
[32]
M. Hu, K. Korschelt, P. Daniel, et al., ACS Appl. Mater. Interfaces 9(2017) 38024-38031.
doi: 10.1021/acsami.7b12212
-
[33]
X. Mu, J. Wang, Y. Li, et al., ACS Nano 13(2019) 1870-1884.
-
[34]
Y. Huang, C. Liu, F. Pu, et al., Chem. Commun. 53(2017) 3082-3085.
doi: 10.1039/C7CC00045F
-
[35]
D. Li, B. Liu, P.J.J. Huang, et al., Chem. Commun. 54(2018) 12519-12522.
doi: 10.1039/C8CC07062H
-
[36]
K. Fan, H. Wang, J. Xi, et al., Chem. Commun. 53(2017) 424-427.
doi: 10.1039/C6CC08542C
-
[37]
D. Jiang, D. Ni, Z.T. Rosenkrans, et al., Chem. Soc. Rev. 48(2019) 3683-3704.
doi: 10.1039/C8CS00718G
-
[38]
M. Liang, X. Yan, Acc. Chem. Res. 52(2019) 2190-2200.
doi: 10.1021/acs.accounts.9b00140
-
[39]
X. Shen, Z. Wang, X. Gao, Y. Zhao, ACS Catal. 10(2020) 12657-12665.
doi: 10.1021/acscatal.0c03426
-
[40]
Y. Ding, G. Wang, F. Sun, Y. Lin, ACS Appl. Mater. Interfaces 10(2018) 32567-32578.
doi: 10.1021/acsami.8b10560
-
[41]
Z. Wang, X. Shen, X. Gao, Y. Zhao, Nanoscale 11(2019) 13289-13299.
doi: 10.1039/C9NR03473K
-
[42]
S. Guo, L. Guo, J. Phys. Chem. C 123(2019) 30318-30334.
doi: 10.1021/acs.jpcc.9b07802
-
[43]
D. Wang, X. Song, P. Li, et al., J. Mater. Chem. B 39(2020) 9028-9034.
-
[44]
X. Gonze, B. Amadon, P.M. Anglade, et al., Comput. Phys. Commun. 180(2009) 2582-2615.
doi: 10.1016/j.cpc.2009.07.007
-
[45]
X. Wang, X.J. Gao, L. Qin, et al., Nat. Commun. 10(2019) 704.
doi: 10.1038/s41467-019-08657-5
-
[46]
S. Guo, L. Guo, J. Phys. Chem. C 123(2019) 30318-30334.
doi: 10.1021/acs.jpcc.9b07802
-
[47]
A.C. Moreno Maldonado, E.L. Winkler, M. Raineri, et al., J. Phys. Chem. C 123(2019) 20617-20627.
doi: 10.1021/acs.jpcc.9b05371
-
[48]
B. Jiang, D. Duan, L. Gao, et al., Nat. Protoc. 13(2018) 1506-1520.
doi: 10.1038/s41596-018-0001-1
-
[49]
Z. Zhang, X. Zhang, B. Liu, J. Liu, J. Am. Chem. Soc. 139(2017) 5412-5419.
doi: 10.1021/jacs.7b00601
-
[50]
S.B. Bankar, M.V. Bule, R.S. Singhal, L. Ananthanarayan, Biotechnol. Adv. 27(2009) 489-501.
doi: 10.1016/j.biotechadv.2009.04.003
-
[51]
R. Ragg, F. Natalio, M.N. Tahir, et al., ACS Nano 8(2014) 5182-5189.
doi: 10.1021/nn501235j
-
[52]
S.G. Rhee, Science 312(2006) 1882-1883.
doi: 10.1126/science.1130481
-
[53]
X. Mei, T. Hu, H. Wang, et al., Biomaterials 258(2020) 120257.
doi: 10.1016/j.biomaterials.2020.120257
-
[54]
M. Comotti, C. Della Pina, R. Matarrese, M. Rossi, Angew. Chem. Int. Ed. 43(2004) 5812-5815.
doi: 10.1002/anie.200460446
-
[55]
M. Comotti, C. Della Pina, E. Falletta, M. Rossi, Adv. Synth. Catal. 348(2006) 313-316.
doi: 10.1002/adsc.200505389
-
[56]
X. Shen, W. Liu, X. Gao, et al., J. Am. Chem. Soc. 137(2015) 15882-15891.
doi: 10.1021/jacs.5b10346
-
[57]
Q. Wang, G. Hong, Y. Liu, et al., RSC Adv. 10(2020) 25209-25213.
doi: 10.1039/D0RA05342B
-
[58]
Y. Yang, D. Zhu, Y. Liu, et al., Nanoscale 12(2020) 13548-13557.
doi: 10.1039/D0NR02800B
-
[59]
F. Charbgoo, M. Bin Ahmad, M. Darroudi, Int. J. Nanomedicine12(2017)1401-1413.
doi: 10.2147/IJN.S124855
-
[60]
J. Yao, Y. Cheng, M. Zhou, et al., Chem. Sci. 9(2018) 2927-2933.
doi: 10.1039/C7SC05476A
-
[61]
S.M. Hirst, A.S. Karakoti, R.D. Tyler, et al., Small 5(2009) 2848-2856.
doi: 10.1002/smll.200901048
-
[62]
T. Pirmohamed, J.M. Dowding, S. Singh, et al., Chem. Commun. (Camb. ) 46(2010) 2736-2738.
doi: 10.1039/b922024k
-
[63]
S. Singh, Biointerphases 11(2016) 04B202.
doi: 10.1116/1.4966535
-
[64]
I. Celardo, J.Z. Pedersen, E. Traversa, L. Ghibelli, Nanoscale 3(2011) 1411-1420.
doi: 10.1039/c0nr00875c
-
[65]
P. Ji, L. Wang, F. Chen, J. Zhang, ChemCatChem 2(2010) 1552-1554.
doi: 10.1002/cctc.201000191
-
[66]
D. Damatov, J.M. Mayer, Chem. Commun. (Camb. ) 52(2016) 10281-10284.
doi: 10.1039/C6CC03790A
-
[67]
V. Nicolini, E. Gambuzzi, G. Malavasi, et al., J. Phys. Chem. B 119(2015) 4009-4019.
-
[68]
I.N. Zelko, T.J. Mariani, R.J. Folz, Free Radical Biol. Med. 33(2002) 337-349.
doi: 10.1016/S0891-5849(02)00905-X
-
[69]
A. Okado-Matsumoto, I. Fridovich, J. Biol. Chem. 276(2001) 38388-38393.
doi: 10.1074/jbc.M105395200
-
[70]
J.M. McCord, M.A. Edeas, Biomed. Pharmacother. 59(2005) 139-142.
doi: 10.1016/j.biopha.2005.03.005
-
[71]
S. Liu, R. Tian, J. Xu, et al., Chem. Commun. 55(2019) 13820-13823.
doi: 10.1039/C9CC07085K
-
[72]
Y. Guan, M. Li, K. Dong, et al., Biomaterials 98(2016) 92-102.
doi: 10.1016/j.biomaterials.2016.05.005
-
[73]
Y. Sheng, I.A. Abreu, D.E. Cabelli, et al., Chem. Rev. 114(2014) 3854-3918.
doi: 10.1021/cr4005296
-
[74]
E.G. Heckert, A.S. Karakoti, S. Seal, W.T. Self, Biomaterials 29(2008) 2705-2709.
doi: 10.1016/j.biomaterials.2008.03.014
-
[75]
G. Calabrese, B. Morgan, J. Riemer, Antioxid. Redox Signal. 27(2017) 1162-1177.
doi: 10.1089/ars.2017.7121
-
[76]
E.V. Kalinina, N.N. Chernov, M.D. Novichkova, Biochemistry (Mosc. ) 79(2014) 1562-1583.
doi: 10.1134/S0006297914130082
-
[77]
N. Couto, J. Wood, J. Barber, Free Radical Biol. Med. 95(2016) 27-42.
doi: 10.1016/j.freeradbiomed.2016.02.028
-
[78]
O.M. Ighodaro, O.A. Akinloye, Alex. J. Med. 54(2018) 287-293.
-
[79]
R. Brigelius-Flohe, M. Maiorino, Biochim. Biophys. Acta 1830(2013) 3289-3303.
doi: 10.1016/j.bbagen.2012.11.020
-
[80]
M.M. Gaschler, A.A. Andia, H. Liu, et al., Nat. Chem. Biol. 14(2018) 507-515.
doi: 10.1038/s41589-018-0031-6
-
[81]
M. Jia, D. Qin, C. Zhao, et al., Nat. Immunol. 21(2020) 727.
doi: 10.1038/s41590-020-0699-0
-
[82]
L.J. Yant, Q.T. Ran, L. Rao, et al., Free Radical Biol. Med. 34(2003) 496-502.
doi: 10.1016/S0891-5849(02)01360-6
-
[83]
Y. Huang, Z. Liu, C. Liu, et al., Chem. Eur. J. 24(2018) 10224-10230.
doi: 10.1002/chem.201801725
-
[84]
T. Wirth, Angew. Chem. Int. Ed. 54(2015) 10074-10076.
doi: 10.1002/anie.201505056
-
[85]
Y. Huang, C. Liu, F. Pu, et al., Chem. Commun. (Camb. ) 53(2017) 3082-3085.
doi: 10.1039/C7CC00045F
-
[86]
N. Singh, M.A. Savanur, S. Srivastava, et al., Angew. Chem. Int. Ed. 56(2017) 14267-14271.
doi: 10.1002/anie.201708573
-
[87]
A.A. Vernekar, D. Sinha, S. Srivastava, et al., Nat. Commun. 5(2014) 5301.
doi: 10.1038/ncomms6301
-
[88]
S. Ghosh, P. Roy, N. Karmodak, et al., Angew. Chem. Int. Ed. 57(2018) 4510-4515.
doi: 10.1002/anie.201800681
-
[89]
B. Yang, Y. Chen, J. Shi, Chem. Rev. 119(2019) 4881-4985.
doi: 10.1021/acs.chemrev.8b00626
-
[90]
V.D. Petrov, F. van Breusegem, AoB Plants (2012) pls014.
-
[91]
H. Sies, Redox Biol. 11(2017) 613-619.
doi: 10.1016/j.redox.2016.12.035
-
[92]
G.P. Bienert, A.L.B. Moller, K.A. Kristiansen, et al., J. Biol. Chem. 282(2007) 1183-1192.
doi: 10.1074/jbc.M603761200
-
[93]
L. Huang, J.X. Chen, L.F. Gan, et al., Sci. Adv. 5(2019) 9.
-
[94]
J. Chen, H. Gao, Z. Li, et al., Chin. Chem. Lett. 31(2020) 1398-1401.
doi: 10.1016/j.cclet.2020.03.052
-
[95]
B. Xu, H. Wang, W. Wang, et al., Angew. Chem. Int. Ed. 58(2019) 4911-4916.
doi: 10.1002/anie.201813994
-
[96]
P. Wang, S. Liu, M. Hu, et al., Adv. Funct. Mater. 30(2020) 2000647.
doi: 10.1002/adfm.202000647
-
[97]
F. Cao, L. Zhang, H. Wang, et al., Angew. Chem. Int. Ed. 58(2019) 16236-16242.
doi: 10.1002/anie.201908289
-
[98]
Y. Tao, E. Ju, J. Ren, X. Qu, Adv. Mater. 27(2015) 1097-1104.
doi: 10.1002/adma.201405105
-
[99]
A. Liu, M. Li, J. Wang, et al., Chin. Chem. Lett. 31(2020) 1133-1136.
doi: 10.1016/j.cclet.2019.10.011
-
[100]
F. Cao, L. Zhang, Y. You, et al., Angew. Chem. Int. Ed. 59(2020) 5108-5115.
doi: 10.1002/anie.201912182
-
[101]
K. Fan, J. Xi, L. Fan, et al., Nat. Commun. 9(2018) 1440.
doi: 10.1038/s41467-018-03903-8
-
[102]
Y. Huang, Z. Liu, C. Liu, et al., Angew. Chem. Int. Ed. 55(2016) 6646-6650.
doi: 10.1002/anie.201600868
-
[103]
J. Shan, X. Li, K. Yang, et al., ACS Nano 13(2019) 13797-13808.
doi: 10.1021/acsnano.9b03868
-
[104]
X. Liu, Z. Yan, Y. Zhang, et al., ACS Nano 13(2019) 5222-5230.
doi: 10.1021/acsnano.8b09501
-
[105]
Z. Wang, Y. Zhang, E. Ju, et al., Nat. Commun. 9(2018) 3334.
doi: 10.1038/s41467-018-05798-x
-
[106]
Q. Wu, Z. He, X. Wang, et al., Nat. Commun. 10(2019) 240.
doi: 10.1038/s41467-018-08234-2
-
[107]
M. Qi, H. Pan, H. Shen, et al., Angew. Chem. Int. Ed. 59(2020) 11748-11753.
doi: 10.1002/anie.202002331
-
[108]
C. Wei, Y. Liu, X. Zhu, et al., Biomaterials 238(2020) 119848.
doi: 10.1016/j.biomaterials.2020.119848
-
[109]
W. Zhen, Y. Liu, W. Wang, et al., Angew. Chem. Int. Ed. 59(2020) 9491-9497.
doi: 10.1002/anie.201916142
-
[110]
Y. Liu, P. Bhattarai, Z. Dai, X. Chen, Chem. Soc. Rev. 48(2019) 2053-2108.
doi: 10.1039/C8CS00618K
-
[111]
L. Minai, D.Y. Hayon, D. Yelin, Sci. Rep. 3(2013) 2146.
doi: 10.1038/srep02146
-
[112]
I.B. Slimen, T. Najar, A. Ghram, et al., Int. J. Hyperthermia 30(2014) 513-523.
doi: 10.3109/02656736.2014.971446
-
[113]
L. Fan, X. Xu, C. Zhu, et al., ACS Appl. Mater. Interfaces 10(2018) 4502-4511.
doi: 10.1021/acsami.7b17916
-
[114]
M. Aioub, S.R. Panikkanvalappil, M.A. El-Sayed, ACS Nano 11(2017) 579-586.
doi: 10.1021/acsnano.6b06651
-
[115]
S.P. Sun, C.J. Li, J.H. Sun, et al., J. Hazard. Mater. 161(2009) 1052-1057.
doi: 10.1016/j.jhazmat.2008.04.080
-
[116]
M. Wang, M. Chang, Q. Chen, et al., Biomaterials 252(2020) 120093.
doi: 10.1016/j.biomaterials.2020.120093
-
[117]
M. Zhou, S. Song, J. Zhao, et al., J. Mater. Chem. B 3(2015) 8939-8948.
doi: 10.1039/C5TB01866H
-
[118]
S. Shen, S. Wang, R. Zheng, et al., Biomaterials 39(2015) 67-74.
doi: 10.1016/j.biomaterials.2014.10.064
-
[119]
S. Li, L. Shang, B. Xu, et al., Angew. Chem. Int. Ed. 58(2019) 12624-12631.
doi: 10.1002/anie.201904751
-
[120]
Y. Jiang, X. Zhao, J. Huang, et al., Nat. Commun. 11(2020) 1857.
doi: 10.1038/s41467-020-15730-x
-
[121]
S. Dong, Y. Dong, T. Jia, et al., Adv. Mater. 32(2020) 2002439.
doi: 10.1002/adma.202002439
-
[122]
A.P. Castano, P. Mroz, M.R. Hamblin, Nat. Rev. Cancer 6(2006) 535-545.
doi: 10.1038/nrc1894
-
[123]
J.P. Celli, B.Q. Spring, I. Rizvi, et al., Chem. Rev. 110(2010) 2795-2838.
doi: 10.1021/cr900300p
-
[124]
M. Wu, Y. Ding, L. Li, Nanoscale 11(2019) 19658-19683.
doi: 10.1039/C9NR06651A
-
[125]
Y. Zhang, F. Wang, C. Liu, et al., ACS Nano 12(2018) 651-661.
doi: 10.1021/acsnano.7b07746
-
[126]
D. Wang, H. Wu, S.Z.F. Phua, et al., Nat. Commun. 11(2020) 357.
doi: 10.1038/s41467-019-14199-7
-
[127]
W. Hiraoka, H. Honda, L.B. Feril Jr., et al., Ultrason. Sonochem. 13(2006) 535-542.
doi: 10.1016/j.ultsonch.2005.10.001
-
[128]
H. Chen, X. Zhou, Y. Gao, et al., Drug Discov. Today 19(2014) 502-509.
doi: 10.1016/j.drudis.2014.01.010
-
[129]
J. Chen, H. Luo, Y. Liu, et al., ACS Nano 11(2017) 12849-12862.
doi: 10.1021/acsnano.7b08225
-
[130]
X. Wang, X. Zhong, F. Gong, et al., Mater. Horizons 7(2020) 2028-2046.
doi: 10.1039/D0MH00613K
-
[131]
F. Gong, L. Cheng, N. Yang, et al., Adv. Mater. 31(2019) 1900730.
doi: 10.1002/adma.201900730
-
[132]
P. Zhu, Y. Chen, J. Shi, ACS Nano 12(2018) 3780-3795.
doi: 10.1021/acsnano.8b00999
-
[133]
X. Zhong, X. Wang, L. Cheng, et al., Adv. Funct. Mater. 30(2019) 1907954.
-
[134]
D. Sun, X. Pang, Y. Cheng, et al., ACS Nano 14(2020) 2063-2076.
doi: 10.1021/acsnano.9b08667
-
[135]
M. Feng, Y. Pan, R. Kong, S. Shu, Innovation (New York, N.Y. ) 1(2020) 100032-100032.
-
[136]
Z. Tang, Y. Liu, M. He, W. Bu, Angew. Chem. Int. Ed. 58(2019) 946-956.
doi: 10.1002/anie.201805664
-
[137]
Z. Tang, P. Zhao, H. Wang, et al., Chem. Rev. 121(2021) 1981-2019.
doi: 10.1021/acs.chemrev.0c00977
-
[138]
Y. Zhu, R. Zhu, Y. Xi, et al., Appl. Catal. B: Environ. 255(2019) 117739.
doi: 10.1016/j.apcatb.2019.05.041
-
[139]
Y.S. Jung, W.T. Lim, J.Y. Park, Y.H. Kim, Environ. Technol. 30(2009) 183-190.
doi: 10.1080/09593330802468848
-
[140]
Y. Sang, F. Cao, W. Li, et al., J. Am. Chem. Soc. 142(2020) 5177-5183.
doi: 10.1021/jacs.9b12873
-
[141]
M. Huo, L. Wang, Y. Chen, J. Shi, Nat. Commun. 8(2017) 357.
doi: 10.1038/s41467-017-00424-8
-
[142]
C. Fang, Z. Deng, G. Cao, et al., Adv. Funct. Mater. 30(2020) 1910085.
doi: 10.1002/adfm.201910085
-
[143]
H. Zhang, X. Liang, L. Han, F. Li, Small 14(2018) 1803256.
doi: 10.1002/smll.201803256
-
[144]
M. Chang, M. Wang, M. Wang, et al., Adv. Mater. 31(2019) e1905271.
doi: 10.1002/adma.201905271
-
[145]
M. Wang, M. Chang, Q. Chen, et al., Biomaterials 252(2020) 120093.
doi: 10.1016/j.biomaterials.2020.120093
-
[146]
B. D'Autreaux, M.B. Toledano, Nat. Rev. Mol. Cell Biol. 8(2007) 813-824.
-
[147]
M.L. Circu, T.Y. Aw, Free Radical Biol. Med. 48(2010) 749-762.
doi: 10.1016/j.freeradbiomed.2009.12.022
-
[148]
N. Singh, M.A. Savanur, S. Srivastava, et al., Nanoscale 11(2019) 3855-3863.
doi: 10.1039/C8NR09397K
-
[149]
Y. Huang, Z. Liu, C. Liu, et al., Angew. Chem. Int. Ed. 55(2016) 6646-6650.
doi: 10.1002/anie.201600868
-
[150]
J. Wu, Y. Yu, Y. Cheng, et al., Angew. Chem. Int. Ed. 60(2020) 1227-1234.
-
[151]
N. Singh, S.K. NaveenKumar, M. Geethika, G. Mugesh, Angew. Chem. Int. Ed. 60(2020) 3121-3130.
-
[152]
S.I. Han, S.W. Lee, M.G. Cho, et al., Adv. Mater. 32(2020) 2001566.
doi: 10.1002/adma.202001566
-
[153]
Y. Liu, Y. Cheng, H. Zhang, et al., Sci. Adv. 6(2020) eabb2695.
doi: 10.1126/sciadv.abb2695
-
[154]
T. Liu, B. Xiao, F. Xiang, et al., Nat. Commun. 11(2020) 2788.
doi: 10.1038/s41467-020-16544-7
-
[155]
X. Mu, J. Wang, Y. Li, et al., ACS Nano 13(2019) 1870-1884.
-
[156]
K. Zhang, M. Tu, W. Gao, et al., Nano Lett. 19(2019) 2812-2823.
doi: 10.1021/acs.nanolett.8b04729
-
[157]
D. Duan, K. Fan, D. Zhang, et al., Biosens. Bioelectron. 74(2015) 134-141.
doi: 10.1016/j.bios.2015.05.025
-
[158]
J. Park, J. Chu, A. Tsou, et al., Biomaterials 32(2011) 3921-3930.
doi: 10.1016/j.biomaterials.2011.02.019

 Login In
Login In





 DownLoad:
DownLoad:
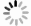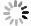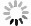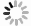
 DownLoad:
DownLoad: